Introduction
Ovarian cancer is a significant global health concern, characterized by high mortality rates and a lack of effective diagnostic tools. The accurate and timely diagnosis of ovarian cancer is crucial for improving patient prognosis and determining appropriate treatment strategies [1]. Medical imaging and tumour visualization are vital steps in the diagnosis of ovarian cancer, and accurate diagnosis using medical imaging is of paramount importance [2]. Medical imaging modalities, such as magnetic resonance imaging (MRI), ultrasound, computed tomography (CT), and positron emission tomography (PET), are commonly used for the diagnosis of ovarian cancer and its staging [3]. However, accurate diagnosis of ovarian cancer using common medical imaging methods can still be challenging, implying the need for more accurate and efficient diagnostic methods.
In recent years, deep learning (DL) has emerged as a promising approach to improve the accuracy of ovarian cancer diagnosis. DL techniques, particularly convolutional neural networks (CNNs), have exhibited remarkable potential in various medical imaging applications. By leveraging the power of artificial intelligence and machine learning algorithms, DL algorithms can automatically extract meaningful features from complex medical images, aiding in the identification and classification of potential pathologies [4,5].
The promise of DL to revolutionize the field of ovarian cancer diagnosis lies in its ability to provide accurate and efficient solutions. Traditionally, the definitive diagnosis of ovarian cancer has relied on histopathological examination, which is time-consuming, labour-intensive, and requires experienced pathologists [6]. Deep learning-based models have the potential to expedite the diagnostic process by quickly recognizing patterns and features in medical images, leading to more efficient and accurate diagnoses [7].
Traditional methods like biomarkers, biopsy, and imaging tests have been the mainstay for ovarian cancer diagnosis, but face limitations including invasiveness, limited early detection accuracy, subjectivity, and inter-observer variability. In contrast, DL leverages advanced neural networks to automatically learn abstract features from raw medical images and data, enabling more objective analysis without manually creating features that are necessary for other machine learning methods [8]. DL uses a non-linear network structure to extract features. It achieves this by combining low-level features, enabling the formation of abstract and complex representations. This approach effectively achieves the essential characteristics of input data, resulting in a distributed representation [9]. Moreover, certain DL approaches have the capability to incorporate a multi-modal fusion framework, enabling the combination of various modalities. This is accomplished by establishing distinct deep feature extraction networks for each modality [10].
Notably, DL has proven extremely promising in the diagnosis of various types of malignancies, including lung cancer [11], thyroid cancer [12], and ovarian cancer [2,13]. Regarding ovarian cancer, DL models have been developed to classify ovarian tissues as malignant, borderline, benign, or normal based on second-harmonic generation imaging, achieving high areas under the receiver operating characteristic curve [13]. Additionally, a preliminary study comparing DL and radiologist assessments for diagnosing ovarian carcinoma using MRI demonstrated that DL exhibited non-inferior diagnostic performance compared to experienced radiologists [2].
The integration of DL into ovarian cancer diagnosis holds immense potential for improving patient outcomes [8,14-16]. This transformative technology can assist clinicians in making more informed decisions, ultimately leading to improved treatment strategies and better patient care. However, further research and validation are necessary to ensure the reliability and generalizability of DL models in routine clinical practice.
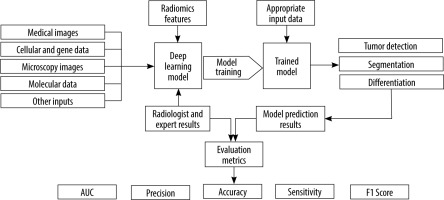
The present study presents a systematic review of the potential and capabilities of DL in improving diagnostic accuracy for ovarian cancer. We will discuss the fundamentals of DL and CNNs, highlighting their applications in ovarian cancer diagnosis. Furthermore, we will review relevant research papers that specifically focus on the use of DL methods in ovarian cancer diagnosis. By examining the existing literature, we aim to gain insights into the current state of DL in this domain and identify potential areas for future research and development. Figure 1 shows an overview of the general workflow of the selected studies.
Material and methods
The research questions guiding this review were twofold: (1) What is the current state of DL in improving the diagnostic accuracy of ovarian cancer using medical imaging? and (2) How does DL contribute to tumour differentiation and radiomics in ovarian cancer diagnosis?
To identify relevant studies, a set of inclusion and exclusion criteria were established. Inclusion criteria encompassed studies published in peer-reviewed journals that focused on DL techniques applied specifically to ovarian cancer diagnosis. The studies were required to involve medical imaging modalities such as MRI, ultrasound, CT, and PET. Additionally, studies evaluating DL approaches for tumour differentiation and radiomics in ovarian cancer diagnosis were included. Exclusion criteria comprised studies not available in English language, those not directly relevant to ovarian cancer or medical imaging, and studies that did not utilize DL techniques.
A comprehensive search strategy was devised by identifying relevant databases, including PubMed, IEEE Xplore, Scopus, and Web of Science. The search query was developed using appropriate keywords and Boolean opera-tors, such as “ovarian cancer”, “deep learning”, “convolutional neural networks”, “medical imaging”, “tomo-graphy”, “MRI”, “ultrasound”, “CT” ,”PET”, “tumor differentiation”, and “radiomics”. The query was refined to ensure accuracy and relevance. Filters were applied to limit the results to publications within the last 6 years. Titles and abstracts of the retrieved articles were thoroughly reviewed to assess their relevance to the research questions and adherence to the inclusion criteria. Full-text versions of potentially relevant articles were obtained for detailed evaluation.
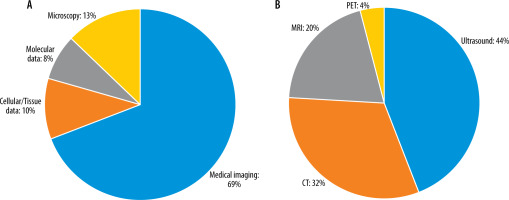
A standardized data extraction form was created to systematically capture pertinent information from each selected study. Details such as study design, sample size, DL techniques employed, medical imaging modalities used, evaluation metrics, and outcomes related to ovarian cancer diagnosis were extracted. The extracted data were meticulously organized to facilitate subsequent analysis and synthesis. Data analysis and synthesis involved summarizing the characteristics and findings of the selected studies. This was accomplished through the creation of a tabular format or narrative synthesis. Common themes, trends, and patterns across the studies were identified and analysed to address the research questions and objectives of the systematic review. Tables 1, 2 show the detailed characteristics of these included studies. Different studies have employed various input types in their chosen networks. Figure 2 illustrates the relative percentage of modalities used as inputs in the selected studies.
Table 1
Participant demographics, algorithm, and model validation for the 39 included studies
| Author [ref.], year | Number of patients | Age (mean, range) (years) | Algorithm | Reference standard |
|---|---|---|---|---|
| Ghoniem et al. [10], 2021 | 587 | NA | Hybrid Deep Learning (CNN – LSTM) | Histopathology |
| Sengupta et al. [17], 2022 | NA | NA | Hybrid Deep Learning (Machine Learning – CNN (Inception Net v3 )) | Histopathology |
| Chen et al. [19], 2022 | 422 | 46.4, NA | ResNet | Histopathology |
| Zhang et al. [9], 2019 | 428 | NA | Hybrid Deep Learning (Machine Learning (uniform local binary pattern) – CNN (GoogLeNet)) | Histopathology |
| Ziyambe et al. [8], 2023 | NA | NA | CNN | Histopathology |
| Mohammed et al. [23], 2021 | NA | NA | Stacking ensemble deep learning model (one-dimensional convolutional neural network (1D-CNN)) | Histopathology |
| Tanabe et al. [24], 2020 | NA | NA | Artificial intelligence (AI)-based comprehensive serum glycopeptide spectra analysis (CSGSA-AI) – CNN (AlexNet) | Histopathology |
| Huttunen et al. [25], 2018 | NA | NA | CNNs (AlexNet, VGG-16, VGG-19, GoogLeNet) | Histopathology |
| Wu et al. [27], 2018 | 85 | NA | CNN (AlexNet) | Histopathology |
| Kilicarslan et al. [33], 2020 | 253 | NA | Hybrid Deep Learning (ReliefF – CNN) | Histopathology |
| Zhao et al. [34], 2022 | 294 | NA | Dual-Scheme Domain-Selected Network (DS2Net) | NA |
| Gao et al. [14], 2022 | 107624 | ≥ 18 | CNN (DenseNet-121) | Histopathology |
| Zhang and Han [35], 2020 | NA | NA | Hybrid Deep Learning (logistic regression classifier (LRC) – CNNs) | Histopathology |
| Shafi and Sharma [37], 2019 | NA | NA | CNN | NA |
| Wang et al. [40], 2022 | 223 | NA | YOLO-OCv2 | NA |
| Zhu et al. [41], 2021 | 100 | NA, 22–45 | CNN | Histopathology |
| Boyanapalli and Shanthini [44], 2023 | NA | NA | Ensemble deep optimized classifier-improved aquila optimization (EDOC-IAO) (ResNet, VGG-16, LeNet) | NA |
| Yao and Zhang [46], 2022 | 224 | NA | CNN (ResNet50) | NA |
| Sadeghi et al. [47], 2023 | 37 | 56.3, 36-83 | 3D CNN (OCDA-Net) | Histopathology – radiologist evaluation |
| Wang et al. [48], 2021 | 451 | 47.8, NA | CNN (ResNet, EfficientNet) | Radiologist evaluation |
| Wang et al. [49], 2023 | 201 | < 30, 30-50, > 50 | CNN (U-net++) | Histopathology |
| Maria et al. [50], 2023 | NA | NA | Hybrid Deep Learning (attention U-Net – YOLO v5) | Radiologist evaluation |
| Christiansen et al. [54], 2021 | 758 | NA | CNN (VGG16, ResNet50, MobileNet) | Histopathology |
| Jung et al. [15], 2022 | 1154 | NA | Convolutional neural network model with a convolutional autoencoder (CNNCAE) (DenseNet, Inception-v3, ResNet) | Histopathology |
| Srilatha et al. [57], 2021 | NA | NA | CNN (U-Net) | NA |
| Saida et al. [2], 2022 | 465 | 50, 20-90 | CNN (Xception) | Histopathology – radiologist evaluation |
| Wang et al. [58], 2021 | 265 | 51, 15-79 | CNN (VGG16, GoogLeNet, ResNet34, MobileNet, DenseNet) | Histopathology |
| Wei et al. [59], 2023 | 479 | 55.61, 19-84 | CNN (ResNet-50) | Histopathology |
| Liu et al. [60], 2022 | 135 | 47, 10-79 | CNN (U-net) | Histopathology |
| Jin et al. [65], 2021 | 127 | 56, 23-80 | U-net, U-net++, U-net with Resnet, CE-Net | Histopathology – radiologist evaluation |
| Jan et al. [66], 2023 | 149 | 46.4, 18-80 | CNN (3D U-Net) | Histopathology – radiologist evaluation |
| Avesani et al. [68], 2022 | 218 | 58, 29-86 | CNN | Histopathology – radiologist evaluation |
| BenTaieb et al. [75], 2017 | 133 | NA | Weakly Supervised Learning Framework | Histopathology |
| Wang et al. [6], 2022 | NA | NA | Hybrid Deep Learning (an weakly supervised cascaded deep learning – a deep learning based classifier) | Histopathology |
| Sundari and Brintha [73], 2022 | NA | NA | Hybrid Deep Learning (intelligent IE with a deep learning-based ovarian tumour diagnosis (IEDL-OVD)) (VGG16 - stacked autoencoder (SAE)) | NA |
| Wang et al. [78], 2018 | 245 | 54.17, NA | Hybrid Deep Learning (DL-CPH) (the deep learning feature – Cox proportional hazard (Cox-PH)) | Histopathology |
| Wang et al. [80], 2022 | 48 | 59.1, 23-79 | Hybrid Deep Learning (ResNet-101 – weakly supervised tumorlike tissue selection model – Inception-v3) | Histopathology |
| Liu et al. [72], 2023 | 185 | 53.33, NA | CNN (ResNet34-CBAM) | Histopathology |
| Ho et al. [83], 2022 | 609 | NA | Deep Interactive Learning (ResNet-182) | Histopathology |
Table 2
Indicator and data source for the 39 included studies
| Author [ref.], year | Number of images (training – testing) | Imaging modality | Source of data | Date range | Open access data |
|---|---|---|---|---|---|
| Ghoniem et al. [10], 2021 | 1375 (1100 train – 275 test) | NA | Retrospective study, data from the TCGA-OV | NA | Yes |
| Sengupta et al. [17], 2022 | NA (75% train – 25% test) | NA | Retrospective study, data from Tata Medical Center (TMC), India | NA | Yes |
| Chen et al. [19], 2022 | 422 (337 train – 85 test) | Ultrasound | Retrospective study, data from Ruijin Hospital and Shanghai Jiaotong University School of Medicine | 2019.01-2019.11 | NA |
| Zhang et al. [9], 2019 | 3843 (3294 train – 549 test) | Ultrasound | Retrospective study, data from database provided at [85] and Peking Union Medical College Hospital | NA | Yes |
| Ziyambe et al. [8], 2023 | 200 (160 train – 40 validation) | Histopathological | Retrospective study, data from the TCGA | NA | Yes |
| Mohammed et al. [23], 2021 | 304 (217 train – 87 test) | NA | Retrospective study, data from Pan-Cancer Atlas | NA | Yes |
| Tanabe et al. [24], 2020 | 351 (210 train – 141 validation) | 2D barcode | Retrospective study, NA | NA | NA |
| Huttunen et al. [25], 2018 | 200 (120 train – 80 validation) | Multiphoton microscopy | Retrospective study, NA | NA | NA |
| Wu et al. [27], 2018 | 1848 (NA) | Cytological | Retrospective study, data from Hospital of Xinjiang Medical University | 2003-2016 | NA |
| Kilicarslan et al. [33], 2020 | 15154 (9092 train – 6062 test, 10608 train – 4546 test, 12123 train – 3031 test) | Gene microarray | Retrospective study, data from public gene microarray datasets | NA | Yes |
| Zhao et al. [34], 2022 | 1639 (1070 train – 569 test) | Ultrasound | Retrospective study, data from Beijing Shijitan Hospital, Capital Medical University | NA | Yes |
| Gao et al. [14], 2022 | 592275 (575930 train – 16345 validation) | Ultrasound | Retrospective study, data from ten hospitals across China | 2003.09-2019.05 | No |
| Zhang and Han [35], 2020 | 1500 (NA) | Ultrasound | Retrospective study, data from stanford.edu/datasets | NA | Yes |
| Shafi and Sharma [37], 2019 | 250 (NA) | MRI | Retrospective study, data from skims (Sher-i-Kashmir Institute of Medical Sciences) and Hospital Kashmir | NA | NA |
| Wang et al. [40], 2022 | 5603 (4830 train – 773 test) | CT | Retrospective study, data from the Affiliated Hospital of Qingdao University | NA | Yes, conditional |
| Zhu et al. [41], 2021 | NA | CT | Retrospective study, data from the Affiliated Hospital of Xiangnan University | 2017.01-2019.01 | Yes |
| Boyanapalli and Shanthini [44], 2023 | NA (70% train – 30% test) | CT | Retrospective study, data from the TCGA-OV | NA | Yes |
| Yao and Zhang [46], 2022 | 224 (157 train – 67 test) | PET/CT | Retrospective study, data from Shengjing Hospital of China Medical University | 2013.04-2019.01 | Yes |
| Sadeghi et al. [47], 2023 | 1224 (1054 train – 170 test) | PET/CT | Retrospective study, data from Kowsar Hospital, Iran | 2019.04-2022.05 | No |
| Wang et al. [48], 2021 | 545 (490 train – 55 test) | MRI | Retrospective study, from one center in the United States | NA | NA |
| Wang et al. [49], 2023 | 201 (152 train – 49 test) | MRI | Retrospective study, data from Hospital Fudan University | 2015.01-2017.12 | Yes, conditional |
| Maria et al. [50], 2023 | 300 (NA) | CT | Retrospective study, data from RSNA and TCIA | NA | Yes, conditional |
| Christiansen et al. [54], 2021 | 3077 (608 train – 150 test) | Ultrasound | Retrospective study, data from the Karolinska University Hospital and Sodersjukhuset in Stockholm, Sweden | 2010-2019 | NA |
| Jung et al. [15] 2022 | 1613 (NA) | Ultrasound | Retrospective study, data from the Seoul St. Mary’s Hospital | 2010.01-2020.03 | Yes, conditional |
| Srilatha et al. [57], 2021 | 150 (NA) | Ultrasound | Retrospective study, from a standard benchmark dataset, available at [86, 87] | NA | Yes |
| Saida et al. [2], 2022 | 3763 (3663 train – 100 test) | MRI | Retrospective study, NA | 2015.01-2020.12 | No |
| Wang et al. [58], 2021 | 279 (195 train – 84 validation) | Ultrasound | Retrospective study, data from Tianjin Medical University Cancer Institute and Hospital | 2013.03-2016.12 | Yes, conditional |
| Wei et al. [59], 2023 | 479 (297 train – 182 validation) | MRI | Retrospective study, data from five center | 2013.01-2022.12 | NA |
| Liu et al. [60], 2022 | 135 (96 train – 39 test) | CT | Retrospective study, data from Nanfang Hospital | 2011.12-2018.08 | Yes, conditional |
| Jin et al. [65], 2021 | 469 (376 train – 93 test) | Ultrasound | Retrospective study, data from Wenzhou Medical University and Shanghai First Maternal and Infant Hospital | 2002.01-2016.12 | Yes, conditional |
| Jan et al. [66], 2023 | 185 (129 train – 56 test) | CT | Retrospective study, data from the MacKay Memorial Hospital | 2018.07-2019.12 | Yes, conditional |
| Avesani et al. [68], 2022 | 218 (152 train – 66 test) | CT | Retrospective study, data from IEO, Milan in Rizzo et al. [88], Fondazione Policlinico Gemelli, Rome; Policlinico Umberto I, Rome; Ospedale Centrale, Bolzano | NA | Yes, conditional |
| BenTaieb et al. [75], 2017 | 133 (68 train – 65 test) | Histopathological | Retrospective study, data from this dataset, available at the following URL: http://tinyurl.com/hn83mvf | NA | Yes |
| Wang et al. [6], 2022 | 288 (187 train – 101 test) | Histopathological | Retrospective study, data from the TCIA, and another dataset, available at https://doi.org/10.7937/tcia.985g-ey35 | NA | Yes |
| Sundari and Brintha [73], 2022 | 216 (151 train – 65 test) | NA | Retrospective study, data from the American National Cancer Institute website | NA | No |
| Wang et al. [78], 2018 | 8917 (5495 train – 3422 test) | CT | Retrospective study, data from the West China Second University Hospital of Sichuan University (WCSUH-SU) and Henan Provincial People’s Hospital (HPPH), available at http://www.radiomics.net.cn/post/111 | 2010.02-2015.09 (WCSUH-SU) – 2012.05-2016.10 (HPPH) | Yes, conditional |
| Wang et al. [80], 2022 | 720 (472 train – 248 test) | Histopathological | Retrospective study, data from Tri-Service General Hospital, National Defense Medical Center, Taipei, Taiwan | 2013.03-2021.01 | Yes, conditional |
| Liu et al. [72], 2023 | 3839 (1909 train – 1930 validation) | MRI | Retrospective study, data from the Affiliated Hospital of Chongqing Medical University | 2013.01-2019.12 | Yes, conditional |
| Ho et al. [83], 2022 | 609 (488 train – 121 test) | Histopathological | Retrospective study, data from Memorial Sloan Kettering Cancer Center | NA | Yes, conditional |
Figure 2
A: Relative percentage of input data types used as network inputs in the study survey. B: Percentage of medical image modalities used as network inputs in the selected studies
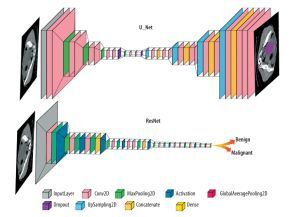
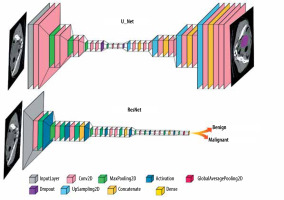
In the reviewed papers, different DL networks were implemented based on their specific objectives. One commonly used architecture is U-Net, which follows an encoder-decoder structure with skip connections and is commonly employed in medical image segmentation tasks. Another widely explored architecture is ResNet, known for its ability to train deep networks using residual blocks, particularly in image classification tasks. The general forms of the ResNet and U-Net architectures are shown in Figure 3, and their detailed configurations and performance metrics are explained in the relevant studies within the results section.
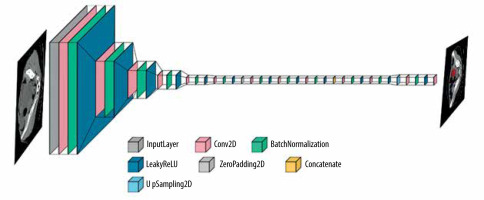
Additionally, for tumour detection, YOLOv5 was identified as a suitable architecture, known for its real-time object detection capabilities. YOLOv5 employs a single-stage detection approach and has demonstrated promising results in detecting and localizing tumours efficiently. Figure 4 shows the layer composition and order in YOLOv5, for efficient tumour detection.
To assess the robustness, performance, and reliability of DL networks for ovarian cancer detection and differentiation, several metrics were commonly used. Accuracy, which measures the overall correctness of DL network predictions, is defined as the ratio of correctly classified samples to the total number of samples in the dataset. Area under the curve (AUC) is a widely used metric that represents the area under the receiver operating characteristic (ROC) curve, indicating the model’s discrimination power. Recall (also known as sensitivity) quantifies the DL network’s ability to correctly identify positive instances, calculated as the ratio of true positives to the total number of actual positives. Precision determines the proportion of correctly predicted positive samples out of all predicted positive samples, while specificity measures the network’s ability to correctly identify negative instances. The F1 score combines precision and recall, providing a balanced evaluation of the network’s performance. Higher values for these metrics generally indicate better performance.
Results
Deep learning techniques in ovarian cancer diagnosis
Ovarian cancer is the fifth most common cause of cancer mortality among women, and early diagnosis is crucial for effective treatment. Deep learning techniques, such as CNNs, have shown potential in improving ovarian cancer diagnosis [10,17]. Deep learning is a subcategory of machine learning that uses artificial neural networks to learn from large amounts of data. Convolutional neural networks are a type of DL algorithm, particularly effective in image recognition tasks. They are composed of multiple layers of interconnected nodes capable of identifying patterns in images and classifying them accordingly [18].
Deep learning techniques have been used to analyse histopathological images of ovarian cancer tissue samples. Algorithms based on DL have the potential to assist in the diagnosis of malignant patterns, predict clinically relevant molecular phenotypes and microsatellite instability, and identify histological features related to patient prognosis and correlated to metastasis.
One study used a multi-modal evolutionary DL model to diagnose ovarian cancer. The model used deep features extracted from histopathological images and genetic modalities to predict the stage of ovarian cancer samples. For processing of the gene data and the histopathological images, an LSTM network and a CNN model were utilized, respectively. Both models were optimized with the Ant Lion Optimization (ALO) algorithm, and finally the outputs of the LSTM and CNN were fused using weighted linear aggregation to make the final prediction. The ReLU activation function, the hyperparameters, and the topology of both networks were automatically selected by ALO. The multi-modal dataset was divided into training, validation, and test sets following a ratio of 6 : 2 : 2, and cross-entropy loss was used for training the model. The model demonstrated remarkable performance with a precision of 98.76%, recall of 98.74%, accuracy of 98.87%, and F1 score of 99.43%. The model was compared to 9 other multi-modal hybrid models and was found to be more precise and accurate in diagnosing ovarian cancer [10].
Another study used a deep hybrid learning pipeline to accurately diagnose ovarian cancer based on nuclear morphology. The images were obtained from ovarian cancer and normal ovary tissues stained for lamin A/B proteins. Extracted features were classified using XGBoost and random forest classifiers and fused with predictions from morphometric parameters of nuclei analysed separately through AdaBoost. A 21-layer CNN architecture was used in this study, and key parameters like filters of 5 × 5, 3 × 3, and 1 × 1 sizes, and leaky ReLU activation were used in the CNN model. To increase training efficiency, controlled data augmentation techniques were applied. The deep hybrid learning model was trained for 250 epochs, with a learning rate of 0.00025 and a batch size of 32. Adam optimization and cross-entropy loss were used during the training process. The model performance was then assessed using the area under the receiver operating characteristic curve (AUC) analysis. The model combined classical machine learning algorithms with vanilla CNNs to achieve a training and validation AUC score of 0.99 and a test AUC score of 1.00 [17].
A retrospective study done by Chen et al. aimed to develop DL algorithms to differentiate benign and malignant ovarian tumours using greyscale and colour Doppler ultrasound images. The ResNet-18 model with ReLU activation function was employed to extract features from each image type. These features were concatenated and then inputted into fully connected layers for classification. The final layer used softmax activation for classification. The performance of the DL algorithms was compared to the OvarianAdnexal Reporting and Data System (O-RADS) and the official ultrasound report. The DL algorithms with feature fusion and decision fusion strategies were developed. The DL algorithm with feature fusion achieved an AUC of 0.93, comparable to the Ovarian-Adnexal Reporting and Data System (O-RADS) and the ultrasound reports. DL algorithms demonstrated sensitivities of 92% and specificities ranging from 80% to 85% for malignancy detection, concluding that DL algorithms based on multimodal US images can effectively differentiate between benign and malignant ovarian tumours, showing comparable performance to expert assessment and O-RADS [19].
In another study, Zhang et al. developed a machine learning-based model to differentiate between benign and malignant ovarian masses/tumours. The mentioned model utilized Google LeNet and a random forest classifier and was able to achieve high diagnostic accuracy for its designed task. A total of 1628 ultrasound images were used, and data augmentation was performed by extracting 9 image patches around the lesion area. The dataset was divided into training (70%), validation (10%), and testing (20%) sets. For evaluation, 10-fold cross-validation was used to calculate performance metrics including accuracy, sensitivity, and specificity. The diagnostic accuracy was 96%, and the sensitivity and specificity were 96% and 92%, respectively [9].
Late-stage diagnosis of ovarian cancer can be challenging due to its vague symptoms and the limitations of current diagnostic methods. Ziyambe et al. introduced a novel CNN algorithm for accurate and efficient ovarian cancer prediction and diagnosis. The CNN consists of 2 conv-ReLU-maxpool blocks, a flattening layer, a fully connected layer, and a softmax layer as output classifier. The CNN model trained on 200 histopathological images augmented to 11,040 images. The optimizer was Adam with a learning rate of 0.001. The training was performed for 50 epochs with a batch size of 32. This model attained a notable accuracy of 94%, correctly identifying 95.12% of malignant cases and accurately classifying 93.02% of healthy cells. This approach may overcome operator-based inaccuracies alongside offering improved accuracy, efficiency, and reliability. Therefore, further research aimed at enhancing the performance of this proposed method is warranted [8].
In a study by Mohammed et al., a stacking ensemble DL model based on a one-dimensional convolutional neural network (1D-CNN) was proposed for multi-class classification sorting of breast, lung, colorectal, thyroid, and ovarian cancers in women using RNASeq gene expression data [20,21]. The model incorporated feature selection using the least absolute shrinkage and selection operator (LASSO) [22]. Each 1D-CNN base model consisted of a single convolutional layer followed by a max pooling layer with a 2 × 2 pooling size. Dense or fully connected layers were then implemented before the final output layer and the softmax activation was used to classify cancer types. The data used in the study was divided into 70% for training, 10% for validation, and 20% for testing purposes. The proposed model showed acceptable results with an accuracy of 93.40%, precision of 73.40%, F1 score of 81.60%, sensitivity of 92%, and specificity of 94.80%. The results demonstrated that the suggested model, with and without LASSO, outperformed other classifiers. Additionally, machine learning methods with under-sampling showed better performance compared to those using oversampling techniques. Statistical analysis confirmed the significant differences in accuracy between various classifiers. The findings suggest that the suggested model can contribute to the early detection and diagnosis of these cancers [23].
Tanabe et al. proposed an artificial intelligence (AI)-based early detection method for ovarian cancer using glycopeptide expression patterns. They converted data on glycopeptide expression in the sera of patients with ovarian cancer and cancer-free patients into 2D barcode information that was analysed by a DL model. They used the pretrained AlexNet model with 25 layers including 5 convolutional and 3 fully connected layers as the base architecture. The last 3 layers were reinitialized and customized for binary ovarian cancer classification. The dataset was divided, with 60% for training and 40% for validation. During training, a batch size of 5, a maximum of 30 epochs, and an initial learning rate of 0.0001 were set. Using their DL model, they were able to achieve a 95% detection rate based on the evaluation of cancer antigen 125 (CA125) and human epididymis protein 4 (HE4) levels [24].
In another study, DL analysis of multiphoton micro-scopy images was used to classify healthy and malignant tissues in reproductive organs. The dataset utilized in this study comprised label-free second harmonic generation (SHG) and 2-photon excited fluorescence (TPEF) images obtained from murine ovarian and reproductive tract tissues. To create binary classifiers, the authors employed 4 pretrained convolutional neural networks (AlexNet, VGG-16, VGG-19, and GoogLeNet). The proposed DL approach achieved a sensitivity of 95% and specificity of 97% in distinguishing healthy tissues from malignant tissues, showcasing its potential as a non-invasive diagnostic tool for ovarian cancer [25].
Computer-aided diagnosis (CADx) can also be used to differentiate between different histologic subtypes of ovarian cancer (mucous, serous, endometroid, and clear cell carcinomas). In a study conducted by Wu Miao et al. an AlexNet [26] based deep convolutional neural network (DCNN) with 5 convolutional layers and 2 fully connected layers was designed to categorize different subtypes of ovarian diseases based on their cytological appearance. Cytological images were used as network input. They prepared the model by 2 gathering input information independently; one was unique picture information and the other was enlarged picture information including picture improvement and picture rotation. The testing results were acquired by the technique for 10-overlap cross-approval, indicating that the exactness of older models improved 72.76-78.20% by utilizing enlarged pictures as preparing information. The created conspire was valuable for grouping ovarian tumours from cytological pictures [27].
Kiliçarslan et al. proposed a hybrid method of utilizing feature ranking and selection (Relief-F) [28] and stacked autoencoder [29,30] approaches for measurement decrease and support vector machines (SVM) [31,32] and CNN for arrangement. The 3 microarray datasets of ovarian, leukaemia, and central nervous system (CNS) were utilized. Among the strategies applied to the 3-micro-array information, the best arrangement precision without measurement decrease was seen with SVM as 96.14% for the ovarian dataset, 94.83% for the leukaemia dataset, and 65% for the CNS dataset. The proposed crossbreed strategy Relief-F + CNN technique beat different methodologies. It gave, respectively, 98.60%, 99.86%, and 80% order exactness for ovarian, leukaemia, and CNS microarray information, individually [33].
Medical imaging modalities for ovarian cancer diagnosis
Ovarian cancer is a challenging disease to diagnose, and medical imaging modalities play an undeniable role in its detection. Different imaging modalities are used in ovarian cancer diagnosis, including MRI, ultrasound, CT, and PET. Each of these modalities has its strengths and limitations in detecting ovarian cancer. For example, MRI is useful in identifying the size and location of ovarian tumours, while CT is better at detecting metastases in the abdomen and pelvis. DL approaches have shown great potential in improving the accuracy of ovarian cancer diagnosis using medical imaging modalities. However, most studies on DL for ovarian cancer diagnosis have focused on CT and MRI modalities.
Zhao et al. proposed a multi-modality ovarian tumour ultrasound (MMOTU) dataset, consisting of 2D ultrasound images accompanied by contrast-enhanced ultrasonography (CEUS) images, with pixel-wise and global-wise annotations. The focus is to perform an unsupervised cross-domain semantic segmentation. To overcome the domain shift issue, they introduced the dual-scheme domain-selected network (DS2Net), which utilizes feature alignment. The proposed DS2Net architecture is a symmetric encoder-decoder structure that employs source and target encoders to extract two-style features from the respective images. Additionally, they introduced the Domain-Distinct Selected Module (DDSM) and the Domain-Universal Selected Module (DUSM) to obtain distinguished and ubiquitous features in each type. These features were fused and fed into source and target decoders to create final predictions. Adversarial losses aligned the feature distributions, while cross-entropy loss was used for segmentation. ReLU activations and SGD/Adam optimizers were employed in this work. To assess segmentation accuracy, evaluation metrics such as intersection-over-union (IoU) and mean IoU were used. The models were trained for 20,000 to 80,000 iterations with a batch size of 1 and an image size of 384 × 384 pixels. Extensive tests and evaluations on the MMOTU dataset demonstrated that DS2Net significantly improves performance for bidirectional cross-domain adaptation of 2D ultrasound and CEUS image segmentation [34].
In another study, Gao et al. aimed to develop a DCNN model based on an automated evaluation of ultrasound images capable of improving existing methods of ovarian cancer diagnosis. The DCNN model utilized a 121-layer DenseNet architecture pretrained on ImageNet. The outputs were adjusted to classify into malignant or benign classes. The network was trained end-to-end using stochastic gradient descent with a momentum of 0.9, weight decay of 0.0001, and a batch size of 32. Data augmentation techniques including resize, rotation, flip, crop, colour jitter, and normalization were employed. Focal loss was employed as the objective function. The resulting DCNN model attained an AUC of 0.91 for the internal dataset, 0.87 for the first external validation dataset, and 0.83 for the second external validation dataset for detecting ovarian cancer. The DCNN model outperformed radiologists in detecting ovarian cancer in the internal dataset and the first external validation dataset. DCNN-assisted diagnosis improved overall accuracy and sensitivity compared to assessment by radiologists alone. The diagnostic accuracy of DCNN-assisted evaluation for 6 radiologists was significantly higher compared to non-assisted evaluation, reaching a total accuracy of 87.6%. Overall, DCNN-enabled ultrasound performed better than the average level of radiologists and matched the expertise of experienced sonologists. It also improved the accuracy of radiologists [14].
Zhang and Han proposed a machine learning algorithm based on a CNN to detect ovarian cancer based on ultrasound images. They created a simple classification network based on US images to detect ovarian cancer. The network consisted of multiple CNNs followed by a U-Net architecture. The CNNs extracted features from the ultrasound images, which were then passed to the U-Net for segmentation and classification. The CNNs comprised convolutional layers for feature extraction, pooling layers for downsampling, and fully connected layers. The U-Net is a typical encoder-decoder with skip connections between the downsampling and upsampling layers. During the training process, a cross-entropy loss function was employed. Sigmoid activations were used in the convolutional layers, while softmax activation was employed in the output layer for classification. They tested the performance of their model, named the Machine Learning-based Convolutional Neural Network-Logistic Regression (ML-CNN-LR) classifier which abstracts obstetric tumour images, based on 2 factors. The precision ratio and recall rate reached 96.50% and 99.10%, respectively [35].
Regarding ultrasounds, Zhang et al. used colour ultrasound images obtained from ovarian masses to predict the probability of early-stage ovarian cancer. Their DL model achieved an overall accuracy of 96% [9].
Shafi and Sharma developed an optimization algorithm named artificial-based colony optimization (ABC-CNN), which identifies the stage of cancer using MRI images. ABC is an optimization algorithm used for feature selection. Characteristics are extracted using kernel PCA algorithms [36] and then classified by the CNN after being trained and tested on MRI images obtained from patients with known cancer stages. Their dataset contained 250 MRI images, with 50 images each for normal, stage 1, 2, 3, and 4 cancers. The dataset. Images were pre-processed by converting to greyscale and applying filters. The highest accuracy achieved was 98.90% [37].
The challenging problem of detecting ovarian tumours on pelvic CT scans using DL is addressed in this study. Wang et al. collected CT images of ovarian cancer patients and developed a new end-to-end network called YOLO-OCv2 (ovarian cancer), based on YOLOv5 [38]. The model employs CSPDarknet53 as the encoder backbone and uses deformable convolutions to capture geometric deformations, a coordinate attention mechanism to enhance object representation, and a decoupled prediction head. For the multitask model, a segmentation head including an ASPP module is added and connected after the PANet feature fusion layers. The networks were trained for 100 epochs using SGD optimizer with initial learning rate of 0.01, momentum of 0.937, and weight decay of 0.0005 on an NVIDIA GPU. The datasets contains 5100 pelvic CT images of 223 ovarian cancer patients, split into training and test sets. The final model achieved the F1 score of 89.85%, mean average precision (mAP) of 74.85% for classification, mean pixel accuracy (MPA) of 92.71%, and mean intersection over union (MIoU) of 89.63% for segmentation. The model incorporates improvements over the previous YOLO-OC [39] model and includes a multi-task model for detection and segmentation tasks [40].
Another study focused on using a CNN algorithm for segmenting ovarian tumours in CT images. The CNN algorithm was applied to perform image segmentation. Their CNN model includes convolutional layers for feature extraction, max pooling for downsampling, and fully connected layers. During the training phase, CT scans of 100 patients with ovarian tumours were used, and the ground truth labels were obtained through manual segmentation performed by doctors. The backpropagation algorithm was used for iterative training to minimize errors between predictions and true labels. Accuracy was evaluated through cross-validation on the dataset. The results showed consistent segmentation outcomes across 3 iterations, with no significant differences in the measured values. The accuracy of the CNN algorithm was 97%, 95%, and 97%, the precision was 98%, 96%, and 98%, and the recall was 96%, 94%, and 96%, respectively, for the 3 measurements, and the 3 F1 scores were 97%, 94%, and 97%, respectively. The CNN algorithm demonstrated high accuracy, precision, recall, and F1 scores, surpassing other segmentation algorithms such as SE-Res Block U-shaped CNN and density peak clustering. Overall, the CNN algorithm proved to be effective in segmenting CT images and could provide reliable guidance for the clinical analysis and diagnosis of ovarian tumours, enhancing their evaluation and facilitating more effective decision-making [41].
Boyanapalli and Shanthini introduced the ensemble deep optimized classifier-improved Aquila optimization (EDOC-IAO) classifier. The proposed EDOC-IAO classifier aimed to overcome the limitations commonly seen in DL methods for early detection of ovarian cancer in CT images. It utilizes a modified Wiener filter [42] for preprocessing, an optimized ensemble classifier (ResNet, VGG-16, and LeNet), and the Aquila optimization algorithm [43] to improve accuracy and reduce over-fitting. Fusion is achieved using average weighted fusion (AWF). The classifier achieved a high accuracy of 96.53% on the cancer genome atlas ovarian (TCGA-OV) dataset, outperforming other methods for ovarian cancer detection [44].
Yao and Zhang developed a model for predicting the malignant vs. benign status of lymph nodes in untreated ovarian cancer patients using PET images. The study included 224 patients with ovarian cancer who underwent PET/CT imaging at a single centre. Image segmentation was performed on the PET images to divide the tumour regions of interest into subregions. They utilized ResNet50 classic architecture without any modification. The model utilizes wavelet transform [45] for feature extraction and image segmentation for parameter evaluation. The model is intended to provide additional information to clinicians and assist in making decisions for first-visit patients. The information was extracted using both image segmentation and wavelet transform. The model was built using a residual neural network and SVM. Results showed that the model achieved high accuracy, AUC, sensitivity, and specificity on the training set (accuracy: 88.54%, AUC: 0.94, sensitivity: 98.65%, specificity: 79.52%) and the test set (accuracy: 91.04%, AUC: 0.92, sensitivity: 81.25%, specificity: 100%). In conclusion, the study displays the effectiveness of wavelet transform and the residual neural network in predicting lymph node metastasis in ovarian cancer patients, providing valuable insights for patient staging and treatment decisions [46].
Also, our study presented the use of 3D CNN for the classification and staging of OC patients using FDG PET/CT examinations. A total of 1224 image sets from OC patients who underwent whole-body FDG PET/CT were investigated and divided into independent training (75%), validation (10%), and testing (15%) subsets. The proposed models, OCDAc-Net and OCDAs-Net, achieved high overall accuracies of 92% and 94% for diagnostic classification and staging, respectively. We introduced the “nResidual” block, which is a variation of the Residual block used in this study. It contains an additional layer on its skip connection, providing an extended structure for feature extraction and learning. Both the Residual and nResidual blocks contribute to the overall architecture of the OCDA-Net and OCDAs-Net models. The models show potential as tools for recurrence/post-therapy surveillance in ovarian cancer [47].
Deep learning for ovarian tumour differentiation
Deep learning has been increasingly used in medical imaging for tumour differentiation, including for ovarian tumours. DL algorithms have shown promising results in accurately differentiating between non-malignant and malignant ovarian tumours using various imaging modalities.
Several studies have evaluated the performance of DL approaches for tumour differentiation in ovarian cancer. For instance, one study developed a DL algorithm that differentiates non-malignant from malignant ovarian lesions by applying a CNN on routine MRI. The study found that the DL model had higher accuracy and specificity than radiologists in assessing the nature of ovarian lesions, with test accuracy of 87%, specificity of 92%, and sensitivity of 75% [48]. Another study aimed to differentiate borderline and malignant ovarian tumours based on routine MRI imaging using a DL model. A total of 201 patients with pathologically confirmed borderline and malignant ovarian tumours were enrolled from a hospital. Both T1- and T2-weighted MR images were reviewed. A U-Net++ model was trained to automatically segment ovarian lesion regions on MR images, and an SE-ResNet-34 model was used for classification. The segmentation-classification framework was designed where the segmented regions were input to a classification model based on CNN to categorize lesions. The study used a DL framework model on a broad dataset of tumour volumes and found that the model was able to identify and categorize ovarian lesions with high accuracy. The DL model computerized network could differentiate borderline from malignant lesions with a significantly higher AUC of 0.87, accuracy of 83.70%, sensitivity of 70%, and specificity of 87.50% based on T2-weighted images [49].
Deep learning is gaining popularity in healthcare, particularly in the detection of ovarian cancer. A hybrid DL approach using YOLOv5 and attention U-Net models has been proposed to accurately detect and segment cancerous ovarian tumours in CT images. YOLO v5 is used for object detection, while Attention U-Net is employed for the segmentation task. The datasets used are from RSNA and TCIA, comprising 300 CT images that have been annotated by a radiologist. These images are resized to 256 × 256, denoised using wavelet transforms, and augmented to create a dataset of 1800 images for training phase. The models are trained end-to-end on this dataset, with hyperparameters such as a learning rate of 0.1 optimized through trial and error. The loss function used is mean squared error for YOLO v5 and binary cross entropy for Attention U-Net. Clinical data has been used to verify the model’s performance, which has shown high accuracy and dice scores. The YOLOv5 model accurately locates ovarian tumours with 98% accuracy, while the attention U-Net model accurately segments the detected tumours with 99.20% accuracy. This computer-aided diagnosis system can assist radiologists in diagnosing ovarian cancer [50].
Christiansen et al. aimed to build a deep neural network and evaluate its capability in distinguishing benign and malignant ovarian lesions using computerized ultrasound image analysis and comparing it to (experts’) subjective assessment [51]. A dataset of ultrasound images acquired from women with known ovarian tumours was categorized based on International Ovarian Tumour Analysis (IOTA) [52,53] criteria by expert ultrasound exa-miners. Transfer learning was applied to 3 pre-trained DNNs, and an ensemble model was created to assess the likelihood of malignancy. Two models were developed: Ovry-Dx1 (benign or malignant) and Ovry-Dx2 (benign, indecisive, or malignant). Diagnostic performance was compared to SA in the test set. Ovry-Dx1 showed similar specificity to SA (86.70% vs. 88%) at a sensitivity of 96%. Ovry-Dx2 achieved a sensitivity of 97.10% and specificity of 93.70%, designating 12.70% of lesions as indecisive. Combining Ovry-Dx2 with SA in indecisive cases did not significantly alter overall sensitivity (96%) and specificity (89.30%) compared to using SA in all cases. Ultrasound image analysis using DNNs demonstrated diagnostic accuracy comparable to human expert examiners in predicting ovarian malignancy [54].
This study aimed to develop a CNN with a convolutional autoencoder (CNN-CAE) model for the accurate classification of ovarian tumours using ultrasound images. A standardized dataset of 1613 ultrasound images from 1154 patients are divided into 5 folds for cross-validation. Ultrasound images were processed and augmented. A CNN with convolutional, max pooling and dense layers, adopting a U-Net architecture with skip connections, was trained to classify images into 5 classes: normal, cystadenoma, mature cystic teratoma, endometrioma, and malignant. The CNN-CAE was optimized using techniques like stochastic gradient descent to minimize the cross-entropy loss between predictions and true labels. The CNN-CAE removes unnecessary information and classifies ovarian masses/tumours into 5 classes. Evaluation metrics include accuracy, sensitivity, specificity, and AUC. Results were verified using gradient-weighted class activation mapping (Grad-CAM) [55]. The CNN-CAE achieved high accuracy, sensitivity, and AUC in normal versus ovarian tumour classification (97.20%, 97.20%, and 0.99, respectively) and distinguishing malignant tumours (90.12%, 86.67%, and 0.94, respectively). The CNN-CAE model serves as a robust and feasible diagnostic tool for accurately classifying ovarian tumours by removing unnecessary data on ultrasound images. It holds valuable potential for clinical applications [15].
Srilatha et al. proposed a CNN-based segmentation model with the combination of GLCM [56] contourlet transformation and CNN feature extraction method. The proposed method contains 4 steps including preprocessing, segmentation, feature extraction, and lastly optimization or classification. These steps were performed on ovarian ultrasound images. The CNN architecture consisted of multiple convolutional layers with kernel sizes ranging from 3 × 3 to 7 × 7, and max pooling layers with pool sizes of 2 × 2. The dataset consisted of 150 ovarian ultrasound images, of which 80 were benign and 70 were malignant. The images were divided into training and test sets. The model achieved accuracy, precision, sensitivity, specificity, and F-score values of 98.11%, 98.89%, 98.75%, 97.06%, and 97.08%, respectively [57].
Another retrospective study by Saida et al. compared the diagnostic performance of DL with radiologists’ interpretations for differentiating ovarian carcinomas from benign lesions using MRI. T2-weighted imaging (T2WI), diffusion-weighted imaging (DWI), apparent diffusion coefficient (ADC) map, and fat-saturated contrast-enhanced T1-weighted imaging (CE-T1WI) were analysed. A CNN was trained on images from patients diagnosed with malignant and non-malignant lesions and evaluated on separate test sets. Images from 194 patients with pathologically confirmed ovarian carcinomas or borderline tumours and 271 patients with non-malignant lesions who underwent MRI were included. Only slices containing the tumour or normal ovaries were extracted and resized to 240 × 240 pixels for the study. The CNN showed sensitivity (77-85%), specificity (77-92%), accuracy (81-87%), and AUC (0.83-0.89) for each sequence, achieving performance equivalent to the radiologists. The ADC map sequence demonstrated the highest diagnostic performance (specificity = 85%, sensitivity = 77%, accuracy = 81%, AUC = 0.89). Deep learning models, particularly when analysing the ADC map, provided a diagnostic performance comparable to experienced radiologists for ovarian carcinoma diagnosis on MRI. These findings highlight the potential of DL in improving the accuracy of ovarian tumour differentiation [2].
Another study intended to assess the capability of a DCNN in distinguishing between benign, borderline, and malignant serous ovarian tumours (SOTs) using ultrasound images. Five different DCNN architectures were trained and tested to perform 2- and 3-class classification on the given images. The datasets consisted of 279 pathology-confirmed SOT ultrasound images from 265 patients. The images were randomly assigned to the training (70%) or validation (30%) sets. Data augmentation techniques such as random cropping, flipping, and resizing were applied to avoid overfitting. The DCNNs were trained for 500 epochs using the Adam optimizer with an initial learning rate of 0.0003 and batch size of 32. Dropout and L2 regularization were used to prevent overfitting. Transfer learning by finetuning the pre-trained ImageNet weights was also explored and compared to training from scratch. Model performance was evaluated by its accuracy, sensitivity, specificity, and AUC. The ResNet-34 model attained the best general performance, with an AUC of 0.96 for distinguishing benign from non-benign tumours and an AUC of 0.91 for predicting malignancy and borderline tumours. The model demonstrated an overall accuracy of 75% for categorizing the 3 SOT categories and a sensitivity of 89% for malignant tumours, outperforming senior sonologists. DCNN analysis of US images shows promise as a complementary diagnostic tool for effectively differentiating between benign, borderline, and malignant SOTs, providing valuable clinical information [58].
Deep learning and radiomics for ovarian cancer diagnosis
Deep learning and radiomics are 2 emerging fields in medical imaging that have shown great potential in improving the diagnostic accuracy of ovarian cancer. Radio-mics refers to the extraction of quantitative characteristics from medical images, which can be used to characterize the tumour phenotype and predict patient outcomes. On the other hand, DL is a subset of machine learning that uses artificial neural networks to learn complex patterns in data. By integrating radiomics and DL, researchers have been able to develop models that can accurately diagnose ovarian cancer and predict patient outcomes.
One study investigated the performance of T2-weighted MRI-based DL and radiomics methods for assessing peritoneal metastases in patients suffering from epithelial ovarian cancer (EOC). The study used ResNet-50 as the architecture of its DL algorithm, and the largest orthogonal slices of the tumour area, radiomics features, and clinical characteristics were used to construct the DL, radiomics, and clinical models, respectively. The ensemble model was then created by combining the 3 mentioned models using decision fusion. The DL model inputs were resized to 224 × 224 voxels, and data augmentation techniques such as brightness/contrast variation, flipping, and rotation were applied to augment the data. The training process utilized the Adam optimizer with a learning rate of 5e-5, a batch size of 64, and a weight decay of 0.01 for 50 epochs. Additionally, one-cycle learning rate scheduling and early stopping techniques were employed. The ensemble model had the best AUCs (0.84, 0.85, 0.87) among all validation sets, outperforming the DL model, radiomics model, and clinical model alone [59].
Ovarian tumours, especially malignant ones, have an overall poor prognosis, making early diagnosis crucial for treatment and patient prognosis. Another study used radiomics and DL features extracted from CT images to establish a classification model for ovarian tumours. This study used a 4-step feature selection algorithm to find the optimal combination of features and then developed a classification model by combining those selected features and SVM. For DL feature extraction, the study designed a binary classification CNN model. The network architecture consisted of modified encoders of U-net added with residual connections, SE-blocks, and fully connected layers. The classification model, which combined radio-mics features with DL features, demonstrated better classification performance with respect to the radiomics features model alone in both the training and test cohort – in the training cohort (AUC 0.92 vs. 0.88, accuracy 89.70% vs. 79.93%), and in the test cohort (AUC 0.90 vs. 0.84, accuracy 82.96% vs. 72.59%) [60].
Jin et al. proposed segmentation algorithms based on U-Net models and evaluated their impact on radiomics features from ultrasound images for ovarian cancer diagnosis. A total of 469 ultrasound images from 127 patients were collected and randomly divided into training (353 images), validation (23 images), and test (93 images) sets. U-net [61], U-Net++ [62], U-Net with Resnet [63], and CE-Net [64] models were used for automatic segmentation. Radiomic features were extracted using PyRadiomics. Accuracy was assessed using the Jaccard similarity coefficient (JSC), dice similarity coefficient (DSC), and average surface distance (ASD), while reliability was evaluated using Pearson correlation and intraclass correlation coefficients (ICC). CE-Net and U-net with Resnet outperformed U-Net and U-Net++ in accuracy, achieving DSC, JSC, and ASD of 0.87, 0.79, and 8.54, and 0.86, 0.78, and 10.00, respectively. CE-Net showed the best radiomics reliability with high Pearson correlation and ICC values. U-Net-based automatic segmentation accurately delineated target volumes on US images for ovarian cancer. Radiomics features from automatic segmentation demonstrated good reproducibility and reliability, suggesting their potential for improved ovarian cancer diagnosis [65].
In a study by Jan et al., an artificial intelligence model combining radiomics and DL criteria obtained from CT images was developed to differentiate between benign and malignant ovarian masses. Various arrangements of feature sets were used to build 5 models, and machine-learning classifiers were used for tumour classification. A total of 185 ovarian tumours from 149 patients were included and divided into training and testing sets. Radio-mics features including histogram, grey-level co-occurrence matrix (GLCM), wavelet, and Laplacian of Gaussian (LoG) features were extracted. In addition, a 3D U-Net convolutional neural network (CNN) was used as a feature extractor from the tumour images. The U-Net consisted of an encoder to extract 224 DL features representing the tumour, and a decoder to reconstruct the original image. The U-Net was trained using an Adam optimizer with a half mean squared error loss function over 25 epochs with a batch size of 1. The best performance was recorded from an ensemble model that combined radiomics, DL, and clinical features, achieving an accuracy of 82%, specificity of 89%, and sensitivity of 68%. The model outperformed junior radiologists in terms of accuracy and specificity and showed comparable sensitivity. The model also improved the performance of junior radiologists when used as an assistive tool, approaching the performance of senior radiologists. The CT-based AI model has the potential to improve ovarian tumour assessment and treatment [66].
Finally, Avesani et al. used multicentric datasets of high-grade serous carcinomas to introduce a model capable of predicting early relapses and breast cancer gene (BRCA) [67] mutation in affected patients. The study utilized pre-operative contrast-enhanced CT scans of the abdomen and pelvis collected from 4 medical centres, totalling 218 patients with high-grade serous ovarian cancer. Manual segmentations of the primary tumour (gross tumour volume) were performed on each CT scan. They developed radiomic models using only handcrafted radiomic features as well as clinical-radiomic models combining radiomic features and relevant clinical variables. Different machine learning algorithms were employed, including penalized logistic regression, random forest, support vector machine (SVM), and XGBoost. Hyperparameter tuning was performed using 5-fold cross-validation. They also employed a 2D convolutional neural network (CNN) for tumour classification. The CNN was trained for 200 epochs with early stopping at 50 epochs of no improvement. Adam optimization and binary cross-entropy loss were used. The proposed model was able to achieve a test AUC of 0.48 for BRCA mutation and 0.50 for relapse prediction. The proposed model predicts handcrafted and deep radiomic features and used the CNN model and give better mutation prediction accuracy [68].
Challenges and opportunities of deep learning in cancer diagnosis
Deep learning has the potential to improve the precision and efficacy of ovarian cancer diagnosis, but there are also challenges and limitations to consider. One of the challenges of DL in ovarian cancer diagnosis is the need for large amounts of high-quality data to train the models effectively. The quality of the data is essential to ensure that the models can generalize to new cases accurately [69]. Utilizing a small dataset during the training phase can result in overfitting [6]. Additionally, DL models may be affected by batch effects when applied to small datasets. In selected studies, there are factors that cause biases that should be addressed. Most of the studies were performed using a limited dataset or single centres that cater to a specific patient demography. Additionally, the data were assessed by a single expert radiologist. To overcome these biases, the authors recommend obtaining data from multiple institutions in the future to introduce more diversity [17,19]. Furthermore, there could be potential biases in the sample selection process, particularly in the selection of cancerous and non-cancerous samples. The authors suggest that revealing clinical details to the person performing tests can ensure unbiased validation in the future [17]. To ensure a comprehensive evaluation of their reliability, it is essential to test the system on larger datasets that encompass various centres, staining techniques, and participant groups [70]. Another challenge is the interpretability of the models. DL models are often considered “black boxes” because it is challenging to understand how they arrive at their conclusions. This can make it difficult to identify and address errors or biases in the models [71]. Another noteworthy point to mention is that in some DL models, the direct correlation between the features extracted by DL and the actual progression of cancer has not been addressed [72]. Most DL approaches have not been utilized for real-time applications in ovarian cancer diagnosis. However, in the future, these approaches have the potential to be employed for real-time diagnostic purposes in ovarian cancer [73].
Despite the challenges, there are several opportunities for further research and development in DL-based diagnostic approaches for ovarian cancer. One opportunity is the use of DL to analyse histopathological images. A recent study proposed a novel CNN algorithm for predicting and diagnosing ovarian malignancies with remarkable accuracy [8]. Another opportunity is the use of DL to analyse other types of data, such as genomic data. DL has shown great potential in analysing genomic data for cancer diagnosis and prognosis [70]. Additionally, DL can be used to develop personalized diagnostic approaches that take into account individual patient characteristics [69].
BenTaieb et al. developed a pathological diagnosis model for ovarian cancer using a histopathological image dataset. Accurate sub-classification of ovarian cancer is crucial for treatment and prognosis. However, pathologists often encounter an inconsistency rate of 13% in classifying ovarian cancer cell types [74]. The final model, based on convolution operations and DL image processing, addresses this issue. It considers differentiated tissue areas as potential variables, disregards nonessential tissue parts, and combines multiple magnified information for accurate diagnosis. The model achieved an average multi-class classification accuracy of 90%. This approach effectively handles intraclass variation in pathological image classification [75].
Despite advancements in surgery and chemotherapy for ovarian cancer, recurrence and drug resistance remain major challenges. Identifying new predictive methods for effective treatment is urgently needed. In another study performed by Wang et al., weakly supervised DL approaches were implemented to assess the therapeutic effect of bevacizumab in patients suffering from ovarian cancer based on histopathologic whole slide images. The model achieved high accuracy, 88.20%; precision, 92.10%; recall, 91.20%; F-measure, 91.70%, outperforming state-of-the-art DL approaches. An independent testing set also yielded promising results. The proposed method can guide treatment decisions by identifying patients who may not respond to further treatments and those with a positive therapeutic response. Statistical analysis using the Cox Proportional Hazards Model [76] showed a significantly higher risk of cancer recurrence for patients predicted to be ineffective compared to those predicted to be effective. Deep learning offers opportunities for predicting therapeutic responses in ovarian cancer patients. The proposed method shows promise in assisting treatment decisions and identifying patients at high risk of cancer recurrence [6].
In another study, Sundari and Brintha designed image enhancement with a deep learning-based ovarian tumour diagnosis (IEDL-OVD) method, which was due to this improved image quality, then improve optimization through the black widow optimization algorithm (BWOA) [77], and used some feature extraction and classification techniques to maximize the precision and recall rate. The IEDL-OVD model has obtained an increased precision and recall of 73.50% and 61.20%, respectively [73].
Based on a study conducted by Wang et al., artificial intelligence can be used to detect and identify multiple prognostic and therapeutic criteria in patients with EOC based on their preoperative medical imaging. In their study, they proposed prognostic biomarkers based on preoperative CT images that can predict the 3-year recurrence probability with decent accuracy (AUC = 0.83, 0.77, and 0.82, in the primary and 2 validation models, respectively). However, larger prospective studies and retrospective validating trials are needed before these can be used in routine clinical practice [78].
In this study, Wang et al. aimed to develop an automated precision oncology framework to find and select EOC and peritoneal serous papillary carcinoma (PSPC) patients who would benefit from bevacizumab treatment. They collected immunohistochemical tissue samples from EOC and PSPC patients treated with bevacizumab and developed DL models for potential biomarkers. The model combined with inflammasome absent in melanoma (AIM2) achieved high accuracy, recall, F-measure, and AUC of 92%, 97%, 93%, and 0.97 in the first experiment, respectively. In the second experiment using cross-validation, the model showed high accuracy, precision, recall, F-measure, and AUC of 86%, 90%, 85%, 87%, and 0.91, respectively. Kaplan-Meier [79] analysis and Cox proportional hazards model confirmed the model’s ability to distinguish patients with positive therapeutic effects from those with disease progression after treatment [80].
Liu et al. developed a deep learning-based signature using preoperative MRI to predict recurrence risk in patients suffering from advanced grade IV serous carcinoma. A hybrid model combining clinical and DL features showed higher consistency and AUC compared to the DL and clinical feature models for 2 validation sets (AUC = 0.98, 0.96 vs. 0.70, 0.67, and 0.50, 0.50). The hybrid model accurately predicted the recurrence risk and 3-year recurrence likelihood, and Kaplan-Meier analysis confirmed its ability to distinguish high and low recurrence risk groups. Based on their study, DL using multi-sequence MRI serves as a low-cost and non-invasive prognostic biomarker for the prediction of high-grade serous ovarian carcinoma (HGSOC) recurrence. The proposed hybrid model eliminates the need for follow-up biomarker assessment by utilizing MRI data [72].
Haematoxylin and eosin (H & E) staining is routinely used to prepare samples for histopathologic assessment. Due to the time-consuming nature of manual annotation for cancer segmentation, DL can be used to automate this labour-extensive process. In that regard. Ho et al. proposed an interactive DL method [81], using a pre-trained segmentation model from another type of cancer (breast cancer) [82], to reduce the burden of manual annotation. Images were first given to the pre-trained model, and mislabelled regions were interactively corrected for training and finetuning the model. The final model achieved an intersection-over-union (IOU) of 0.74, recall of 86%, and precision of 84%. The proposed approach was able to achieve accurate ovarian cancer segmentation with minimal manual annotation time. High-grade serous ovarian cancer patches were used to train the model. In conclusion, deep interactive learning offers a solution to reduce manual annotation time in cancer diagnosis, enhancing efficiency and the potential for classification of cancer subtypes [83].
Discussion
The use of DL methods, particularly CNNs has demonstrated significant potential in improving the diagnostic accuracy of ovarian cancer. Studies have reported high accuracy, sensitivity, and specificity of these techniques in distinguishing between benign and malignant ovarian tumours [2,9,17]. In fact, DL models trained on various medical imaging modalities have outperformed traditional methods and even surpassed the performance of expert radiologists in detecting ovarian cancer [2,84]. The successful implementation of DL algorithms on histopathological images and clinical imaging modalities, such as ultrasound, CT, MRI, and PET images, has shown promising results in identifying cancerous cells and tumours, predicting the stage of the disease, and performing tumour segmentation. These advancements in DL have the potential to significantly impact early detection and diagnosis of ovarian cancer, leading to improved patient outcomes and more effective decision-making in clinical practice.
Despite the promising results, there are still several areas in the field of DL for ovarian cancer diagnosis that require further research and exploration. A comparative analysis with traditional diagnostic methods or other prevalent DL models within the field remains absent. Such a comparative assessment would provide a comprehensive understanding of the efficacy and limitations of DL models in contrast to conventional approaches. It would also offer insights into the relative advantages and challenges posed by DL techniques, enabling a more nuanced interpretation of their potential clinical impact and feasibility in real-world scenarios.
Factors such as interpretability, data heterogeneity, and model generalizability across diverse populations and healthcare settings warrant detailed examination. Furthermore, elucidating the challenges in integrating clinical data, genetic information, and multi-modal imaging for more robust predictive models is essential. Addressing these limitations can guide researchers toward overcoming barriers and refining DL methodologies for improved clinical applicability. Acknowledging these limitations can contribute to a more nuanced understanding of the scope and applicability of DL models in real-world clinical settings.
It is recommended that larger-scale studies be conducted using diverse and representative datasets to validate the performance of DL models across different populations [10]. Additionally, exploring the potential of DL in other imaging modalities, such as PET and molecular imaging, could lead to further improvements in the detection and diagnosis of ovarian cancer [3]. Combining multiple modalities or hybrid models should also be investigated to leverage the complementary strengths of different imaging techniques in diagnosing ovarian cancer effectively. Standardized protocols and benchmarks should be developed to assess the performance of DL models in ovarian cancer diagnosis, ensuring consistent and reliable results [10]. The generalizability and transferability of DL models across different healthcare settings and institutions should also be assessed. Integrating clinical data, genetic information, and other relevant factors into DL models could enhance their predictive capabilities and personalized medicine approaches. Furthermore, addressing the interpretability and explainability of DL models is crucial to gain trust and acceptance from clinicians and patients.
Conclusions
The use of DL methods, particularly CNNs, has shown great promise in enhancing the diagnostic accuracy of ovarian cancer. These techniques have demonstrated high accuracy, sensitivity, and specificity in distinguishing between benign and malignant ovarian tumours, surpassing the performance of traditional methods and expert radiologists. By applying DL algorithms to various medical imaging modalities, such as ultrasound, CT, MRI, and PET images, significant progress has been made in identifying cancerous cells, predicting disease stages, and performing tumour segmentation. Nevertheless, there are still important research directions to pursue in the field of DL for ovarian cancer diagnosis. Cost-effectiveness analyses should be conducted to evaluate the economic impact of implementing DL techniques in routine clinical practice for ovarian cancer diagnosis [50,69]. By addressing these research areas, further advancements can be made in leveraging DL techniques to enhance the accuracy, efficiency, and reliability of ovarian cancer diagnosis. This will eventually lead to better patient prognosis and more informed healthcare decision-making.