Introduction
The development of deep learning (DL), an emerging field of artificial intelligence (AI), has greatly facilitated clinical decision support for interpreting medical images such as echocardiograms, chest radiographs, and magnetic resonance images (MRI) [1,2]. The use of DL algorithms to diagnose internal joint derangement through MRI analysis presents numerous possibilities. Investigational DL algorithms for internal joint derangement have been developed to detect tears in the anterior cruciate ligament (ACL), meniscus, and articular cartilage in the knee, rotator cuff tears in the shoulder, Achilles tendon tears in the ankle, as well as to identify nerves, bones, and muscles [1,2]. Pre-vious DL-based knee MRI studies have mostly focused on a single area such as the ACL, meniscus, or articular cartilage [3-14]. Recent studies have used 2-dimensional (2D) and 3-dimensional (3D) convolutional neural network (CNN) DL models and MRI data to developed automated algorithms that detect and grade abnormalities of multiple joint tissues [15,16]. Despite these advances, inputting image datasets remains a challenge for complex DL algorithms.
Fat suppression technique is crucial for visualizing oedema, cartilage structures, and bone marrow lesions. Sagittal fat-suppressed proton-density or T2-weighted images have shown higher detection rates for abnormalities in the ligaments, bone marrow, articular cartilage, meniscus, and soft tissues [17,18]. In this study, we hypothesized that abnormal findings could be more easily detected by subtracting knee fat-suppression images without abnormal findings from those with abnormal findings using DL. To test our hypothesis, we developed a DL model with 2D CNNs to accurately generate fat-suppression images from original non-fat-suppression images acquired with 2 different sequences, T1-weighted imaging (T1WI), and intermediate-weighted imaging, as well as to generate fat-suppression images in which abnormal findings were completely removed (referred to as knee normal fat-suppression images). We then used the generated images to detect and classify abnormalities using the fat-suppression image-subtraction method, which involved subtracting abnormal minus normal images. The purpose of the study is to develop a DL model that can accurately generate fat-suppression images and easily detect and classify abnormalities on knee MRI.
Material and methods
This study was conducted with the approval of the Ethics Committee in our institution (S20064). The use of clinical data for this research was disclosed on the institutional website, and the potential participants were given the opportunity to decline to be further enrolled in the study.
MRI acquisition
All images were obtained in our institution using a 3 T MR scanner (Magnetom Skyra, Siemens Healthcare, Erlan-gen, Germany) with an 8-channel knee coil. All studies consisted of 2D-FSE T1-weighted (T1WI) and intermediate-weighted images, with and without fat suppression in the sagittal plane. The parameters are shown in Table 1. All the images were extracted in Digital Imaging and Communications in Medicine (DICOM) file format, converted to 8-bit greyscale Portable Network Graphics (PNG) format, and resized to 128 × 128 pixels.
Sample selection
Forty-five knee studies in 45 consecutive symptomatic patients (mean age 54.6 ± 20.3 years; 16 males/29 females; 21 right/24 left) performed at 3 T in our institution between April 2020 and July 2020 were included. Cases after ligament reconstruction were excluded. The final diagnoses were osteoarthritis (n = 18), meniscal tear (n = 38), ligament tear (n = 5), post-resection of benign tumour (n = 2), Osgood-Schlatter disease (n = 1), and muscle injury (n = 1). In addition, 12 knee MR studies in 6 healthy volunteers who had neither symptoms nor history of trauma in the knee (mean age 34.2 ± 9.5 years; 4 males/2 females; 6 right/6 left) were included.
Deep learning (DL) model
The DL model uses 2D CNNs on the open-source Neural Network Console ver.2.1 deep learning library (A. Hayakawa et al., unpublished data, 2021), which was commercially developed (Sony Network Communications, Tokyo, Japan, https://dl.sony.com) and was based on the Python programming language (version 3.6.3; Python Software Foundation, Wilmington, DE, USA), running on a computer (eX.computer, Windows 10 operating system) with an AMD Ryzen 9 3950X 3.5 GHz processor, 64 GB RAM, and an NVIDIA GeForce RTX 2080Ti 11 GB graphics processing unit (NVIDIA, Santa Clara CA, USA).
Our DL algorithm consisted of two consecutive processes: generation of fat-suppression images and detection and classification of abnormalities (Figure 1).
Figure 1
Structure of deep learning model. A) Encoder-decoder network (Endeco-Net) for normal fat-suppression images. B) U-Net for fat-suppression images incorporating both normal and abnormal findings. C) Convolutional neural network for subtracting the generated normal fat-suppression images (Figure 1A) from the generated fat-suppression images with abnormal findings (Figure 1B) and detecting and classifying abnormal findings. D) Inception module in the convolutional neural network (Figure 1C)

Generation of fat-suppression images
In this process, we developed 2 types of DL models using 2D CNNs: one for generating fat-suppression images without any abnormal findings (containing only normal findings), and another for generating fat-suppression images with normal and/or abnormal findings. The first DL model was specifically designed to synthesize fat-suppression images without any abnormal findings (referred to as normal fat-suppression images) using an in-house convolutional encoder-decoder network (Endeco-Net) (Figure 1A). For this model, as a control group, we exclusively used normal MR images from 12 knee studies involving healthy volunteers. Consequently, the Endeco-Net was trained solely on normal findings and was capable of generating only normal findings. Thus, we created 348 image datasets, each consisting of one slice of original T1WI, original intermediate-weighted images, original fat-suppressed (FS)-intermediate-weighted images, and generated FS-intermediate-weighted images at the same cross-section.
Next, we developed a DL model utilizing a U-Net network for faithfully synthesizing fat-suppression images incorporating both normal and/or abnormal findings (Figure 1B). By using the 45 abnormal knee MR studies from 45 patients, we created 1263 image datasets, each consisting of one slice of original T1WI, original intermediate-weighted images, original FS-intermediate-weighted images, and generated FS-intermediate-weighted images at the same cross-section.
Detection and classification of abnormalities on knee MRI
In this process, apart from the previously mentioned Endeco-Net and U-Net, we developed a dedicated DL model for subtracting the generated normal fat-suppression images (FS-intermediate-weighted images) (fat-suppression images without any abnormal findings) from the generated fat-suppression images (FS-intermediate-weighted images) with abnormal findings in order to effectively detect abnormalities. Furthermore, we developed an additional DL model specifically designed for the detection and classification of these abnormalities (Figures 1C and D).
Based on the luminance of the subtraction images, a threshold of 128 out of 256 colour tones between free fluid and oedema was determined. Thus, the colour blue was displayed when a certain area was free fluid (≥ 128), while the colour red was displayed when it was judged to be oedema (≤ 128). The value of 128 was employed to effectively exclude free fluid. Prior to setting the value of 128, a radiologist assessed whether it was possible to distinguish between free fluid and oedema in several cases. Therefore, abnormalities of the ACL, bone marrow, articular cartilage, and menisci are depicted in the colour red. Attenuation map images were generated by superimposing the subtraction images on acquired intermediate-weighted images.
We augmented the image data by randomly zooming in and out, rotating within a range of –0.15 and +0.15 radians, and flipping it left and right [19]. We primarily focused on fine structures in the knee joints, so we applied such data augmentation.
From the 1263 image datasets, each consisting of one slice of original T1WI, original intermediate-weighted images, and original FS-intermediate-weighted images, a total of 2472 image datasets, each consisting of one slice of original T1WI, original intermediate-weighted images, FS-intermediate-weighted images synthesized using Endeco-Net, FS-intermediate-weighted images synthesized using U-Net, and subtraction images between the FS-intermediate-weighted images synthesized using Endeco-Net and synthesized using U-Net at the same cross-section, were created.
Image assessments
Two board-certified radiologists independently assessed the image quality of the 2472 FS-intermediate-weighted images synthesized by Endeco-Net, 2472 FS-intermediate-weighted images synthesized by U-Net, and 2472 subtraction images between these fat-suppression images. One of the radiologists conducted a second evaluation after a one-month interval. The image assessments were performed on a liquid crystal display monitor (dia-gonal 80 cm [31.5”], screen ratio 16:9; resolution 2560 × 1440, 3.7 megapixels). Regarding inter-reader agreement by 2 radiologists and intra-reader agreement by one radiologist, kappa values were calculated. The strength of agreement quantified by the kappa statistic was graded as follows: < 0, poor; 0.01-0.20, slight; 0.21-0.40, fair; 0.41-0.60, moderate; 0.61-0.80, substantial; and 0.81–1, almost perfect [20].
In regard to the labelling of image sets, we labelled each image set. The presence or absence of overall abnormalities on the image sets, which consisted of one slice each of the original sagittal T1WI, original sagittal intermediate-weighted images, and original sagittal FS-intermediate-weighted images at the same cross-sections, was determined by the radiologist. In addition, the presence or absence of abnormalities involving the ACL, bone marrow, articular cartilage, and menisci, joint effusion with capsular distention, soft-tissue oedema, and other fluid collections was also assessed. Thus, determinations of abnormal findings were made on a per-slice basis. In cases where equivocal findings were encountered, the adjacent 1- or 2-slice images were reviewed to reach a definitive conclusion. Finally, a cross-check was conducted with clinical diagnoses based on the medical information.
“ACL abnormality” was defined as a complete tear, partial tear, or mucoid degeneration of the ACL. A partial tear of the ACL was defined as any fibre discontinuity of up to 80%. Mucoid degeneration of the ACL was a thickened, ill-defined ACL, with increased signal intensity on all MR sequences but without fibre discontinuity [8]. Because these categories are difficult to distinguish even on the radiologist’s readings of MRI, we did not differentiate among them. “Bone marrow abnormality” was defined as any bone marrow oedema pattern. “Cartilage abnormality” was defined as articular cartilage irregularity, focal defect, or diffuse thinning due to cartilage degeneration at the medial and lateral femorotibial joints and patellofemoral joints [6]. “Meniscus abnormality” was defined as a vertical tear, horizontal tear, complex tear, irregular-shaped meniscus, or disappeared or displaced meniscus.
Statistical analysis
We divided the image datasets, including the FS-intermediate-weighted images synthesized by U-Net, and the subtraction images, into 3 groups for training, validation, and testing. To evaluate our DL model, 5 metrics of predictive power (accuracy, average precision, average recall, F-measure, and sensitivity) were calculated on a Neural Network Console ver. 2.1 deep learning library (Sony). In addition, area under the receiver operator characteristic curve (AUROC) values were also calculated with commercial software (SPSS for Windows ver. 28.0, IBM, Armonk, NY, USA).
Results
Generation of fat-suppression images
Out of the 2472 image datasets (each consisting of one slice of original T1WI, original intermediate-weighted images, FS-intermediate-weighted images synthesized using Endeco-Net, FS-intermediate-weighted images synthesized using U-Net, and subtraction images between the FS intermediate-weighted images synthesized using Endeco-Net and synthesized using U-Net at the same cross-section), 2203 (89.1%) were deemed to have adequate image quality, while 269 (10.9%) were determined to have inadequate image quality. In terms of the breakdown of judging of inadequate image quality, the fat suppression images synthesized through the Endeco-Net and U-Net were considered satisfactory. However, 10.9% of the subtraction images synthesized by Endeco-Net and U-Net were judged to have inadequate image quality. One radiologist determined that the 10.9% of the subtraction images did not affect the diagnosis of internal derangements in the knee joint.
The inter-reader agreement was substantial (k value: 0.77), and the intra-reader agreement was almost perfect (k value: 0.88).
Detection and classification of abnormalities on knee MRI
The image datasets for training, validation, and testing were 1799 (81.7%), 99 (4.5%), and 305 (13.8%), respectively. A summary of the training, validation, and testing image datasets is shown in Table 2.
Table 2
A summary of datasets for training, validation, and testing
Of the 2203 image datasets, 976 (44.3%) were interpreted by the radiologist as “normal” and 1227 (55.7%) were determined to be “abnormal”. Of the “abnormal” images, an ACL abnormality was detected in 11.9%, a bone marrow abnormality in 27.3%, a cartilage abnormality in 55.0%, a meniscus abnormality in 64.9%, and other diagnoses in 14.4%.
Accuracy, average precision, average recall, F-measure, and sensitivity of our DL model for determining whether presence or absence of overall abnormalities on knee MRI were 89.5%, 89.4%, 89.4%, 89.4%, and 90.5%, respectively. The AUROC (95% confidence interval [CI]) was 0.931 (0.899–0.963). Accuracies, average precisions, average recalls, F-measures, sensitivities, and AUROCs to detect each abnormality are shown in Table 3.
Table 3
Results of metrics to detect abnormalities in knee MR imaging by DL model with 2D CNNs
[i] DL – deep learning, 2D CNN – 2-dimensional convolutional neural network, AUROC – area under receiver operating characteristic curve, 95% CI – 95% confidence interval, ACL – anterior cruciate ligament. Others include joint effusion with capsular distention, soft-tissue oedema, and soft-tissue fluid collection. *Overall means presence or absence of any abnormalities on knee MR images
Representative cases are shown in Figures 2 and 3.
Figure 2
A 53-year-old male. (A) Original sagittal T1-weighted image, (B) original sagittal intermediate-weighted image, (C) original sagittal fat-suppressed intermediateweighted image, (D) fat-suppressed intermediate-weighted image synthesized by U-Net, (E) fat-suppressed intermediate-weighted image synthesized by Endeco-Net (normal fat-suppression image), (F) subtraction image between fat-suppression images synthesized by U-Net and synthesized by Endeco-Net, (G) attenuation map image. (D, E) Fat-suppression intermediate-weighted images synthesized by U-Net and Endeco-Net both are of adequate image quality. (D, F, G) Joint effusion (arrow), bone marrow edema pattern and cartilage loss (thin arrow), and meniscus tear (curved arrow) are shown on the fat-suppressed intermediate-weighted image synthesized by U-Net, subtraction image, and attenuation map image. Joint effusion is represented in blue color, while bone marrow edema pattern and cartilage loss and meniscus tear are shown in red color on the attenuation map. Our deep learning model detected bone marrow abnormality, cartilage abnormality, and meniscus abnormality
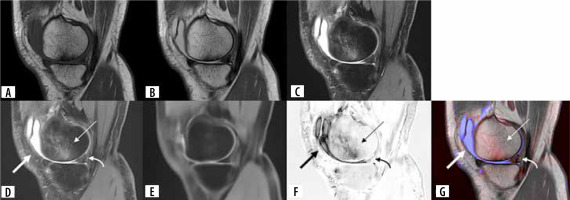
Figure 3
A 63-year-old male. (A) Original sagittal T1-weighted image, (B) original sagittal intermediate-weighted image, (C) original sagittal fat-suppressed intermediate-weighted image, (D) fat-suppressed intermediate-weighted image synthesized by U-Net, (E) fat-suppressed intermediate-weighted image synthesized by Endeco-Net (normal fat-suppression image), (F) subtraction image between fat-suppression images synthesized by U-Net and by Endeco-Net, (G) attenuation map image. (D, E) Fat-suppression intermediate-weighted images synthesized by U-Net and Endeco-Net both are of adequate quality. (D, F, G) Joint effusion (arrow) and anterior cruciate ligament abnormality (thin arrow) are shown on the fat-suppressed intermediate-weighted image synthesized by U-Net, subtraction image, and attenuation map image. Joint effusion is represented in blue color. A red color overlay is observed on the ACL, which indicate an ACL abnormality on the attenuation map. Our deep learning model detected anterior cruciate ligament abnormality
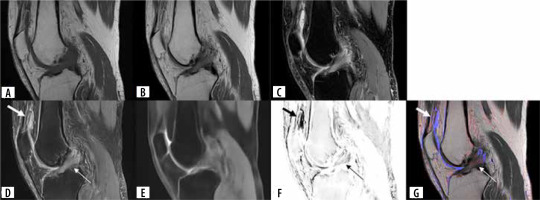
Discussion
We developed a DL model with 2D CNNs for the synthesis of fat-suppression images from 2 different non-fat-suppressed 2D-FSE sequences (T1WI and intermediate-weighted-image) and presented a fat-suppression subtraction-image method using 2D CNN DL algorithms for the detection and classification of abnormal findings on knee MRI. The accuracy, average precision, average recall, F-measure, and sensitivity of our DL model for determining the presence or absence of overall abnormalities on knee MRI were 89.5%, 89.4%, 89.4%, 89.4%, and 90.5%, respectively. The AUROC was 0.931. For the specific abnormalities, the sensitivity was 57.4% for bone marrow oedema, while for ACL, cartilage, meniscus, and others it ranged from 75.0% to 80.0%. The accuracy was between 89.5% and 95.1% for all abnormalities. The AUROC was between 0.910 and 0.979. These results indicate that our DL model with 2D CNNs can generate fat-suppression images of sufficient quality from 2 different non-fat-suppressed 2D-FSE sequences and detect and classify abnormalities on knee MRI through the fat-suppression image-subtraction method.
Fayad et al. [21], who developed a DL model utilizing 2D CNNs for generating FS-intermediate-weighted-images from non-FS-intermediate-weighted-images with 3D-FSE, demonstrated the feasibility of DL-based synthesis of high-quality fat suppression images. In our study, 10.9% of the image datasets were determined to have inadequate image quality. Because there are no previous studies for comparison, it is challenging to determine whether this percentage is high or low. This 10.9% specifically pertained to issues observed in the subtraction images between the fat-suppression images synthesized through Endeco-Net and U-Net. Indeed, the fat-suppression images synthesized by Endeco-Net and U-Net were found to be satisfactory. The reasons for inadequate image quality were identified as blurring and misregistration at the anatomical edges, particularly in the medial and lateral aspects of the knee, observed in the subtraction images. This may be due to the process of the synthesis of fat-suppression images without any abnormality. For the removal of any abnormal findings, we used an encoder-decoder to perform data reduction. In addition, only one sequence series of an original 2D-FSE intermediate-weighted imaging dataset was used as an input at the training. Initially, we considered including 2 sequence series as an input for Endeco-Net; however, we found that a lower data volume of input was more effective in removing abnormal findings and in generating fat-suppression images without any abnormal findings (referred to as normal fat-suppression images). This aspect represents one of the key principles of our DL models. While, for the synthesis of fat-suppression images with normal and/or abnormal findings, a U-Net network was used. In addition, original 2D-FSE T1WI and intermediate-weighted images were used as an input. This decision was made based on the belief that at least 2 different imaging sequences were necessary to accurately synthesize fat-suppression images with abnormalities. In any case, the sagittal plane images of the medial and lateral aspects of the knee joint, comprising only skin and subcutaneous fat, generally have minimal impact on the diagnosis of knee joint derangements. One radiologist determined that the 10.9% of subtraction images did not affect the diagnosis of internal derangements in the knee joint.
Previously, Bien et al. [15] developed a DL model using 2D CNNs for detecting abnormalities in the knee (MRNet) using a dataset of 1370 knee MRIs, including all 3 orthogonal planes. The reference standards were a majority vote of 3 musculoskeletal radiologists. Their DL model achieved accuracy, sensitivity, and AUROC of 85%, 87.9%, and 0.937 for detecting overall abnormality on knee MRI, respectively. In the following 2 studies, there are ambiguous aspects in the definition of general/overall abnormality. Irmakci et al. [22], who attempted to evaluate several DL models for abnormality detection in the knee, reported varying accuracy ranging from 82.5% to 85.8%, sensitivity ranging from 96.8% to 97.9%, and AUROC ranging from 0.811 to 0.909. Tsai et al. [23] reported accuracy of 91.7%, sensitivity of 96.8%, and AUROC of 0.941. In our study, the accuracy, sensitivity, and AUROC of our DL algorithms for the detection of overall abnormalities were 89.5%, 90.5%, and 0.931, respectively. The AUROC in our series was nearly on par with their results, while the accuracy and sensitivity were superior to those of Bien’s study.
For the specific abnormalities, our results showed high accuracy (95.1%) and AUROC (0.979) but a low sensitivity of 75% for ACL tears. Bien et al. [15] reported a sensitivity of 76% for ACL tears, although the specificity was 97%, while Chang et al. [6] demonstrated a sensitivity of 100% and an accuracy of > 96%. Liu et al. [24] used a 2D CNN DL model to investigate another ACL assessment approach for binary ACL-tear classification and reported 96% for both sensitivity and specificity and 0.980 for an AUROC. Germann et al. [7] reported a DL model using 3D CNNs for detecting ACL tears with a sensitivity of 96.1%, a specificity of 93.1%, and an AUROC of 0.935. Irmarci et al. [22] reported sensitivity of 77.8%, specificity of 93.9%, accuracy of 86.7%, and AUROC of 0.954. Zhang et al., [11] using 3D CNN model reported sensitivity of 97.6%, specificity of 94.4%, accuracy of 95.7%, and AUROC of 0.960. Astuto et al. [16], using a 3D CNN DL model and 3D-FSE MRI datasets, reported sensitivity of 88% and specificity of 89% for the detection of ACL abnormalities. In our study, we designed a DL model using 2D CNNs. The accuracy for the detection of ACL abnormalities in our series was comparable to these previous studies. The sensitivity for the detection of ACL abnormalities was similar to that in Bien’s study [15], but both sensitivities were lower than the other previous studies. Namiri et al. [3] previously reported that 2D and 3D CNN DL models performed similarly in classifying ACL abnormalities. They reported sensitivity of 76.4% and 82.4% and specificity of 93.7% and 99.6%, respectively. Recently, Shin et al. [25], who used a 2D CNN DL model and one oblique-sagittal image along the ACL on which the largest ACL area was observed, reported accuracy of 94.1% and AUROC of 0.941. Our series included only a small number of ACL abnormalities. Although it is well known that most ACL tears are visible on sagittal images, [17] the entire ACL was not shown on a single-slice sagittal image. Therefore, for the detection of ACL abnormalities, cropping the images to the ACL, multi-slice input, or multi-plane input might increase the sensitivity.
Regarding the cartilage abnormalities, our results showed that the accuracy, sensitivity, and AUROC were 89.8%, 80%, and 0.947, respectively. Liu et al. [5] reported the sensitivity ranging from 80.5% to 84.1%, specificity from 85.2% to 87.9%, and AUROC from 0.914 to 0.917 using 2D CNN and 2D FSE FS-T2WI in the sagittal plane. Astuto et al. [16], using 3D CNN and 3D FSE images, reported sensitivity of 85%, specificity of 89%, and AUROC of 0.930. Our results were quite similar to theirs. In meniscus abnormalities, a relatively large number of studies using DL have previously been reported [4,9, 12,15,16,24-27]. Multi-slice or multi-plane input might increase the sensitivity in the detection of meniscus tears. Liu et al. [5] also found that DL-based detection had substantial intraobserver agreement, but clinical radiologists had moderate to substantial inter-observer agreement. DL was less prone to errors due to inexperience, distraction, or fatigue but had a high false-positive rate and was not effective in evaluating images with disparate parameters.
To date, only a limited number of studies have employed DL for the detection of bone marrow abnormalities. Astuto et al. [16], using DL for the detection through 3D CNN and 3D FSE sequences, reported lower sensitivity (70%) for the detection of bone marrow oedema compared to other tissues. Fayed et al. [21], who developed a DL model utilizing 2D CNNs for generating FS-intermediate-weighted-images from non-FS-intermediate-weighted-images with 3D-FSE, reported sensitivity of 76% and specificity of 90% when human readers interpreted the generated FS-intermediate-weighted-images. Kijowski et al. [28] reported the same trend in their evaluation for bone marrow oedema on 3D FSE FS-intermediate-weighted images by 2 musculoskeletal radiologists, with lower sensitivity (85.3%). In our study, although using DL detection based on 2D sequences, the accuracy and AUROC for the detection of bone marrow abnormalities were 89.5% and 0.910, respectively. However, the sensitivity was relatively low at 57.4%. This suggests that it is difficult to determine if bone marrow findings are abnormal or not due to significant variations in the size, location, and signal intensity of bone marrow abnormalities in the femur, tibia, and patella, even though our DL models were able to identify changes in bone marrow signals. We think that establishing a gold standard for bone marrow abnormality is also challenging. As a future step, to improve the diagnostic accuracy of bone marrow abnormality, it may be necessary to augment the number of cases and improve training data for the DL models. Additionally, incorporating the bone marrow finding with other findings could promote a comprehensive assessment.
A summary of DL studies for abnormal detection on knee MRI is presented in Table 4 [3-7,9,11-13,15,16,22-27,29].
Table 4
A summary of DL studies for abnormality detection on knee MRI
| Author | Year | Analysed pulse sequence | Field strengths [T] | DL model | Sensitivity | Specificity | Accuracy | AUROC | Comments |
|---|---|---|---|---|---|---|---|---|---|
| Overall abnormality | |||||||||
| Bien et al. [15] | 2018 | 2D*, sag FST2, cor T1, ax PD | 1.5, 3 | 2D CNN | 87.9% | 71.4% | 85% | 0.937 | |
| Irmakci et al. [22] | 2020 | 2D*, sag T2, cor T1, ax PD | 1.5, 3 | 2D CNN | 96.8-97.9% | 28-40% | 82.5-85.8% | 0.811-0.909 | The definition of general abnormality is unclear. |
| Tsai et al. [23] | 2020 | 1.5, 3 | 2D CNN | 96.8% | 72% | 91.7% | 0.941 | Analysed pulse sequences are unclear. The definition of general abnormality is unclear. | |
| Range | 87.9-97.9% | 28-72% | 82.5-91.7% | 0.811-0.941 | |||||
| The present study | 2D*, sag T1, IWI | 3 | 2D CNN | 90.5% | 89.5% | 0.931 | |||
| Anterior cruciate ligament abnormality | |||||||||
| Bien et al. [15] | 2018 | 2D*, sag FST2, cor T1, ax PD | 1.5, 3 | 2D CNN | 75.9% | 96.8% | 86.7% | 0.965 | |
| Chang et al. [6] | 2019 | 2D*, cor PD | 1.5, 3 | 2D CNN | 100% | 93.3% | 96.7% | Mucoid degeneration and partial tear are excluded. | |
| Liu et al. [24] | 2019 | 2D*, sag FST2, sag PD | 3 | 2D CNN | 96% | 96% | 0.980 | ||
| Germann et al. [7] | 2020 | 2D*, cor STIR, sag FST2 | 1.5, 3 | 3D CNN | 96.1% | 93.1% | 0.935 | ||
| Irmakci et al. [22] | 2020 | 2D*, sag T2, cor T1, ax PD | 1.5, 3 | 2D CNN | 77.8% | 93.9% | 86.7% | 0.954 | |
| Zhang et al. [11] | 2020 | 2D*, sag FSPD | 1.5, 3 | 3D CNN | 97.6% | 94.4% | 95.7% | 0.960 | |
| Namiri et al. [3] | 2020 | 3D*, sag FSPD | 3 | 2D CNN | 82.4% | 93.7% | Full-thickness tears are assessed. | ||
| Namiri et al. [3] | 2020 | 3D*, sag FSPD | 3 | 3D CNN | 76.4% | 99.6% | Full-thickness tears are assessed. | ||
| Tsai et al. [23] | 2020 | 1.5, 3 | 2D CNN | 92.3% | 89.1% | 90.4% | 0.960 | Analysed pulse sequences are unclear. | |
| Awan et al. [13] | 2021 | 2D*, sag FSPD | 1.5 | 2D CNN | 91.7% | 94.7% | 0.980 | ||
| Astuto et al. [16] | 2021 | 3D*, sag FSPD | 3 | 3D CNN | 88% | 89% | 0.900 | ||
| Shin et al. [25] | 2022 | 2D*, sag FST2 | 1.5 | 2D CNN | 94.1% | 0.941 | Single oblique-sagittal images along the anterior cruciate ligament are assessed. | ||
| Tran et al. [29] | 2022 | 2D*, 3D*; sag, cor FSPD/FST2 | 1, 1.5, 3 | 2D CNN | 87% | 91% | 90.2% | 0.941 | MRI studies from 12 imaging centres are included. |
| Range | 75.9-100% | 89-99.6% | 86.7-96.7% | 0.900-0.980 | |||||
| The present study | 2D*, sag T1, IWI | 3 | 2D CNN | 75% | 95.1% | 0.979 | |||
| Bone marrow abnormality | |||||||||
| Astuto et al. [16] | 2021 | 3D*, sag FSPD | 3 | 3D CNN | 70% | 88% | 0.830 | ||
| Range | 70% | 88% | 0.830 | ||||||
| The present study | 2D*, sag T1, IWI | 3 | 2D CNN | 57.4% | 89.5% | 0.910 | |||
| Cartilage abnormality** | |||||||||
| Liu et al. [5] | 2018 | 2D*, sag FST2 | 3 | 2D CNN | 80.5-84.1% | 85.2-87.9% | 0.914-0.917 | ||
| Pedoia et al. [4] | 2019 | 3D*, sag FSPD | 3 | 3D CNN | 80% | 80.3% | 0.880 | Patellar cartilages are assessed. | |
| Astuto et al. [16] | 2021 | 3D*, sag FSPD | 3 | 3D CNN | 85% | 89% | 0.930 | ||
| Range | 80-85% | 80.3-89% | 0.880-0.930 | ||||||
| The present study | 2D*, sag T1, IWI | 3 | 2D CNN | 80% | 89.8% | 0.947 | |||
| Meniscus abnormality | |||||||||
| Bien et al. [15] | 2018 | 2D*, sag FST2, cor T1, ax PD | 1.5, 3 | 2D CNN | 71% | 74.1% | 72.5% | 0.847 | |
| Pedoia et al. [4] | 2019 | 3D*, sag FSPD | 3 | 3D CNN | 82% | 89.8% | 0.890 | ||
| Fritz et al. [12] | 2020 | 2D*, sag FSIWI, cor STIR | 1.5, 3 | 3D CNN | 91.2% | 87.1% | 90% | 0.961 | |
| Irmakci et al. [22] | 2020 | 2D*, sag T2, cor T1, ax PD | 1.5, 3 | 2D CNN | 61.5-69.2% | 76.5-85.3% | 70.0-75.8% | 0.779-0.808 | |
| Tsai et al. [23] | 2020 | 1.5, 3 | 2D CNN | 86% | 89% | 88% | 0.904 | Analysed pulse sequences are unclear. | |
| Rizk et al. [9] | 2021 | Sag, cor FSPD | 1, 1.5, 3 | 3D CNN | 67-89% | 84-88% | 82-87% | 0.840-0.930 | Analysed pulse sequences are unclear. Medial and lateral meniscus are separately assessed. |
| Astuto et al. [16] | 2021 | 3D*, sag FSPD | 3 | 3D CNN | 85% | 85% | 0.930 | ||
| Li et al. [26] | 2022 | 2D*, sag FSPD | 3 | 3D CNN | 94.1% | 78.5% | 92.4% | 0.907 | |
| Shin et al. [27] | 2022 | 2D*. sag, cor FST2 | 1.5 | 2D CNN | 78.6% | 93.3% | 92.0% | 0.924 | |
| Range | 61.5-94.1% | 74.1-93.3% | 70-92.4% | 0.779-0.961 | |||||
| The present study | 2D*, sag T1, IWI | 3 | 2D CNN | 79.4% | 89.5% | 0.943 | |||
DL – deep learning, AUROC – area under the receiver operator characteristic curve, 2D* – 2-dimensional pulse sequence, 3D* – 3-dimensional pulse sequence, sag – sagittal image, cor – coronal image, axi – axial image, FS – fat-suppressed, T2 – T2-weighted image, T1 – T1-weighted image, PD – proton density-weighted image, IWI – intermediated-weighted image, 2D – two-dimensional, 3D – three-dimensional, CNN – convolutional neural network
There are several limitations to this study. Firstly, the number of image data included in this study, acquired from both patients and healthy volunteers, was small, and the knee MRI protocols and parameters were fixed. External cross-validation is necessary to confirm our preliminary observations [30]. Larger studies in an uncontrolled environment are also needed to assess the clinical usefulness of this method. Secondly, we did not use a surgical standard of reference for correlation. Thirdly, in this study, we employed a value of 128 within a range of 256 colour tones to differentiate between free fluid and oedema, especially aiming to enhance the specificity of bone marrow abnormality detection. Before setting the value of 128, a radiologist deliberated if it was possible to distinguish between free fluid and oedema in several cases. Because there might be overlap between free fluid and oedema, reassessment of the threshold might be required. Fourthly, due to being based on the accuracy on a per-slice basis, there appears to be a tendency for the diagnostic accuracy to be relatively low. Moreover, in cases where the ACL, articular cartilage, and meniscus are entirely absent within a slice, free fluid may be detected. However, typically, even if oedema is not detected at the site where these tissues are absent, it is common for findings of oedema to be present in the neighbouring tissues. Alternatively, in consecutive adjacent slices, a finding of oedema is detected, which is expected to be present in the surrounding tissues. Hence, 3-dimensional analysis may be considered necessary to improve the diagnostic accuracies. Finally, we did not evaluate the diagnostic performance of human readers when assisted by our DL model. Although our initial results are promising, further technical development and correlation with surgical findings as the gold standard will be required before this method can be implemented fully in clinical practice.
Conclusions
We present a DL model that can generate fat suppression images of sufficient quality from 2 different non-fat-suppression images and detect and classify abnormalities on knee MRI. This method could be useful even in cases of poor-quality fat suppression images or when fat suppression images are unavailable. Furthermore, our results suggest that the use of a DL model with 2D CNNs for fat-suppression subtraction-image method could be a useful tool in detecting and classifying abnormalities on knee MRI.


